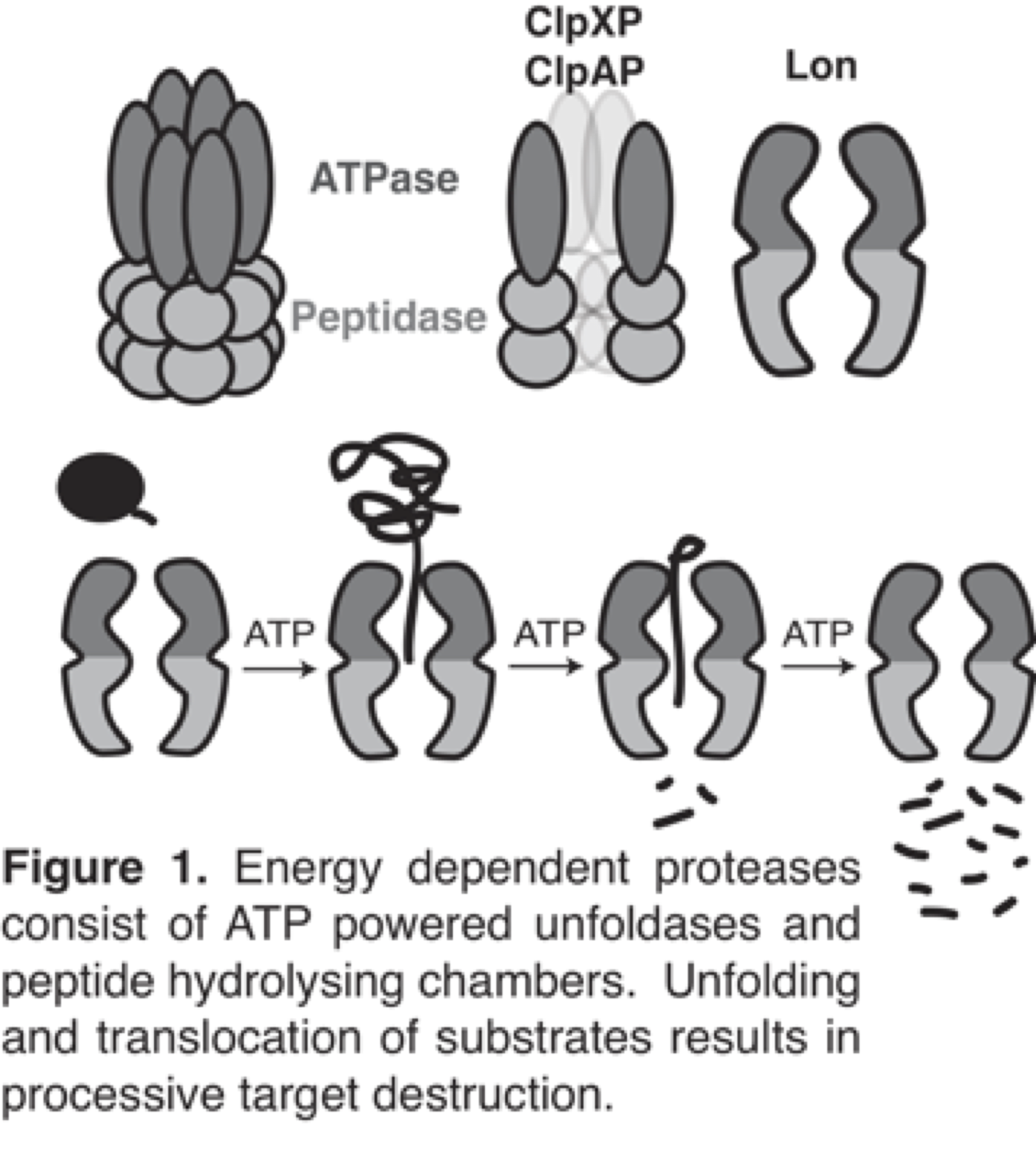
Protein degradation is an essential process for all biological life. Damaged or improperly folded proteins need to be cleared from the cell before they elicit toxic effects. Regulatory proteins need to be degraded so that the response they support exists only as long as it is necessary. However, as proteolysis is an irreversible event, great care must be taken to only degrade those factors as needed without disturbing the balance of other proteins. Degradation of proteins in bacteria is controlled by energy dependent proteases. These oligomeric proteases comprise unfoldase and peptidase activities encoded on separate proteins (e.g. the ClpXP and ClpAP proteases) or in separate domains within the same polypeptide (e.g., the Lon protease).The unfoldase component captures ATP hydrolysis to recognize, unfold and translocate target proteins to the peptidase compartment where the protein is hydrolyzed. Although proteases alone degrade some targets, adaptor proteins can further expand or alter substrate choice. Because these proteases have both regulatory and housekeeping roles, it is vital to understand how proteases maintain specificity while accommodating a wide range of substrates.
DRIVING THE BACTERIA CELL-CYCLE WITH REGULATED PROTEIN DEGRADATION
The α-proteobacterium Caulobacter crescentus undergoes an ordered series of growth and division known as the cell-cycle. As in eukaryotes, this cell-cycle is driven by waves of phosphorylation and protein degradation. This regulated degradation requires the ClpXP protease, although ClpXP levels are unchanged during the cell-cycle, indicating that it must be selectively activated. We are addressing this selective activation using purified proteins to reveal new insights into the mechanism and control of this complex process.
Identifying critical protease substrates.
Although the ClpXP protease is essential in Caulobacter no substrates had been identified that were toxic to cells when not degraded supporting the existence of additional unknown substrates. We used an epitope-tagged inactive ClpP to capture >100 putative ClpXP substrates in vivo that covered all major biological functions, and validated a set of these biochemically and in vivo. One substrate was the transcription factor TacA, stabilization of which results in mistiming of morphological changes, directly integrating protease activity with bacterial development. We also identified a number of replication factors, such as DnaX, as potential ClpXP substrates. This was particularly satisfying as loss of ClpX is toxic in this species and we have now shown that processing of replication factors is one of the species specific features of ClpXP in Caulobacter that is needed to sustain viability.
Defining an adaptor hierarchy for cell-cycle dependent proteolysis.
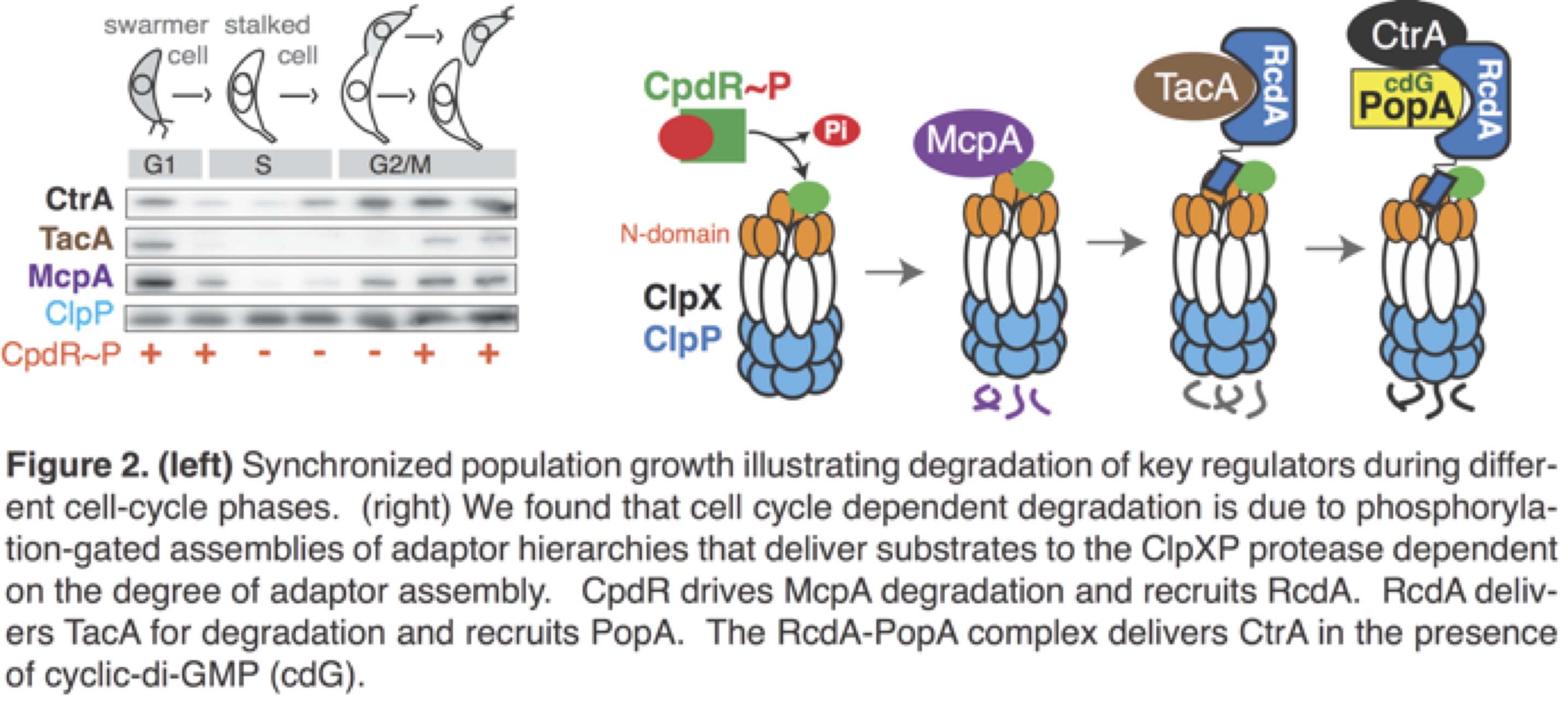
Based on the proteomic work above and additional experiment by others, it is clear that the ClpXP protease degrades many proteins during the cell cycle, including the essential transcription factor CtrA, the developmental regulator TacA, the phosphodiesterase PdeA, and the chemoreceptor McpA. Interestingly, despite the cyclic degradation of their substrates, levels of both ClpX and ClpP remain constant. Therefore, there must be additional regulation of the ClpXP protease that sustains cell cycle degradation.
Although ClpXP is active on its own, the proteins CpdR, RcdA and PopA are required for cell-cycle dependent protein degradation, but their role was unclear prior to our work. In 2015 we reported that cell cycle dependent phosphorylation of CpdR gates its ability to prime the ClpXP protease for degradation of one class of substrates (e.g. McpA, PdeA). Once primed, the protease can now recruit the adaptor RcdA, which delivers a second class of substrates (including TacA) for destruction. Finally, triggered by an increase in cyclic di-GMP levels, PopA is recruited to form an RcdA-PopA complex that destroys a third class of substrates (CtrA). We are currently identifying additional substrates that require specific degrees of adaptor assembly, structurally characterizing adaptors, and determining how adaptors function in other bacteria.
CONTROLLING DNA REPLICATION THROUGH STRESS-RELATED PROTEASES
Orderly growth requires that cells respond to environmental stress, and regulated protein degradation actively contributes to these responses. Replication of DNA is arguably the most regulated molecular event for life. We are currently exploring new ways that stress responses, proteolysis and replication converge to control cell survival.
Partial proteolysis produces essential clamp loader diversity.
Chromosome replication requires ring-shaped clamps that are dynamically assembled on DNA by loader complexes. In bacteria, the DnaX subunit of the clamp loader complex exists as two forms: a longer form (tau) and a shorter form (gamma). In E. coli, gamma is generated by programmed ribosomal frameshifting during translation of the dnaX transcript. This frameshift site is poorly conserved among dnaX orthologs, leading to the speculation that g formation is not universal. Surprisingly, we found that partial proteolysis of DnaX by ClpXP generates gamma in Caulobacter. Both forms of DnaX are essential for normal growth and processing dynamics contribute to DNA damage tolerance. Thus, partial proteolysis may be a widely conserved mechanism in bacteria to generate multiple functions from a single gene. How proteases stall and the role of these conserved DnaX forms in DNA replication are among our ongoing goals for this project.
Proteolytic control of replication during stress.
Upon proteotoxic stress, Caulobacter growth is arrested because the replication initiator DnaA is rapidly degraded. In a collaborative work, we discovered that misfolded proteins resulting from proteotoxic conditions can allosterically activate the Lon protease, directly connecting products of proteotoxic stress and regulatory proteolysis . We are investigating how misfolded proteins and other regulators such as DNA allosterically activate Lon. This aspect is particularly exciting, as Lon was originally identified as a DNA-binding protein, the biological significance of which is still unknown. Our ability to reconstitute these activities, combined with our detailed knowledge of Lon’s role in Caulobacter affords us a perfect opportunity to bridge this gap with the goal of identifying conserved functions for DNA-binding by Lon. Most recently we have used systems-level approaches such as RNA-seq, tn-seq and proteomics to determine the impact of proteases across a range of stresses.
We are always interested in having engaging, enthusiastic scientists of all levels in the lab. If you are interested in joining the Chien lab, please email Peter: pchien@umass.edu.